Chapter 4: Geocentric Models
library(rethinking)
library(tidyverse)
library(splines)
library(gridGraphics)
library(cowplot)Easy
4E1. \(y_i \sim Normal(\mu,\sigma)\) defines the likelihood.
4E2. There are two parameters in the posterior distribution (\(\mu\) and \(\sigma\)).
4E3. \(P(\mu,\sigma|y) = \frac{\prod_i Normal(y_i|\mu, \sigma) Normal(\mu| 0, 10)Exp(\sigma|1)}{\int\int\prod_iNormal(y_1|\mu, \sigma)Normal(\mu|178,20)Exp(\sigma|1)d\mu d\sigma}\)
4E4. \(\mu_i = \alpha + \beta x_i\) is the linear model in the model definition.
4E5. There are three parameters in the posterior distribution (\(\alpha\), \(\beta\), \(sigma\)).
Medium
4M1. Simulate the prior for y.
#number of simulations to draw
n <- 1e4
#simulate the prior for mu
mu <- rnorm(n, mean = 0, sd = 10)
#simulate the prior for sigma
sigma <- rexp(n, rate = 1)
#simulate y using the simulated values for mu and sigma
prior_y <- rnorm(n, mean = mu, sd = sigma)
#plot the simulated prior
dens(prior_y)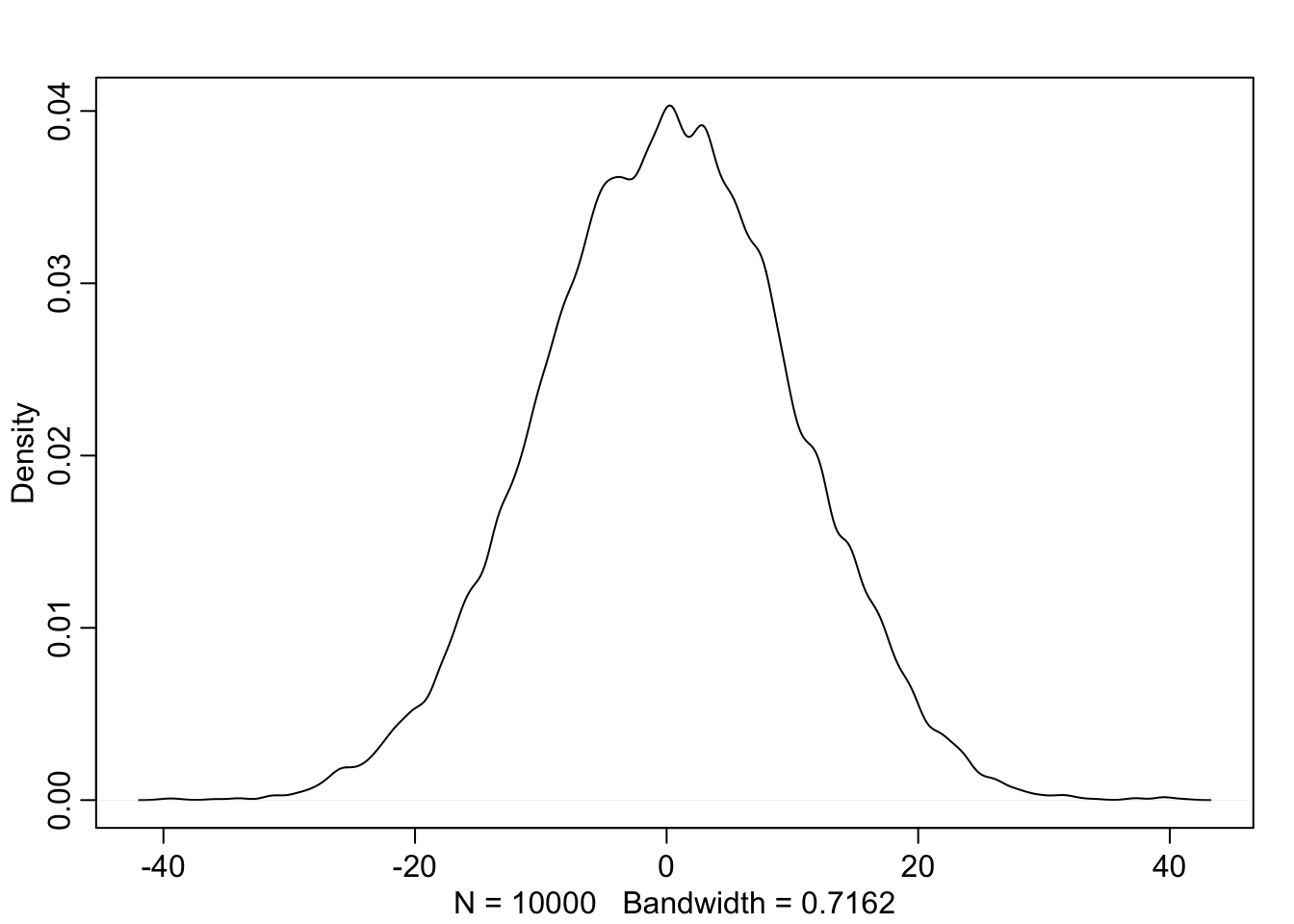
4M2.
quap(
alist(
y ~ dnorm(mu, sigma),
mu ~ dnorm(0, 10),
sigma ~ dexp(1)
))4M3.
\[\displaylines{ y_i \sim Normal(\mu, \sigma) \\ \mu_i = \alpha + \beta X \\ \alpha \sim Normal(0, 10) \\ \beta \sim Uniform(0, 1) \\ \sigma \sim Exponential(1))} \]
4M4. Choosing Priors
#load in Howell dataset
data("Howell1")
#subset data to only look at kids
kids <-
Howell1 %>%
filter(age <= 18)
#get mean and standard deviation for priors of alpha
round(mean(kids$height),0)## [1] 110round(sd(kids$height),0)## [1] 26#kids age 10
kids_10<-
kids %>%
filter(age == 10)
#kids age 16
kids_16 <-
kids %>%
filter(age == 16)
#height increase per year between ages 10 and 16 assuming a steady rate
round((mean(kids_16$height) - mean(kids_10$height))/6,0)## [1] 4The Mathematical Model \[\displaylines{ h_i \sim Normal(\mu, \sigma)\\ \mu_i = \alpha + \beta Y \\ \alpha \sim Normal(110, 26)\\ \beta \sim Normal(4, 1.5) \\ \sigma \sim Uniform(0, 50) }\]
4M5. Assuming that height never decreases as age increases,I would change the prior on \(\beta\) from \(\beta \sim Normal(4, 1.5)\) to \(\beta \sim Log-Normal(4, 1.5)\) as a log-normal distribution enforces positive numbers.
4M6. If the variance (\(\sigma^2\)) among any age group doesn’t exceed 64cm, I would change the prior on \(\sigma\) from \(\sigma \sim Uniform(0,50)\) to \(\sigma \sim Uniform(0, \sqrt{64})\).
4M7.
adults <-
Howell1 %>%
filter(age >= 18)m4.3_nomean <- quap(
alist(
height ~ dnorm( mu, sigma),
mu <- a + b*weight,
a ~ dnorm(178, 20),
b ~ dlnorm(0, 1),
sigma ~ dunif(0, 50)),
data = adults)round(vcov(m4.3),3)## a b sigma
## a 0.073 0.000 0.000
## b 0.000 0.002 0.000
## sigma 0.000 0.000 0.037round(vcov(m4.3_nomean),3)## a b sigma
## a 3.601 -0.078 0.009
## b -0.078 0.002 0.000
## sigma 0.009 0.000 0.037There’s much more covariance when the weight isn’t centered by the mean.
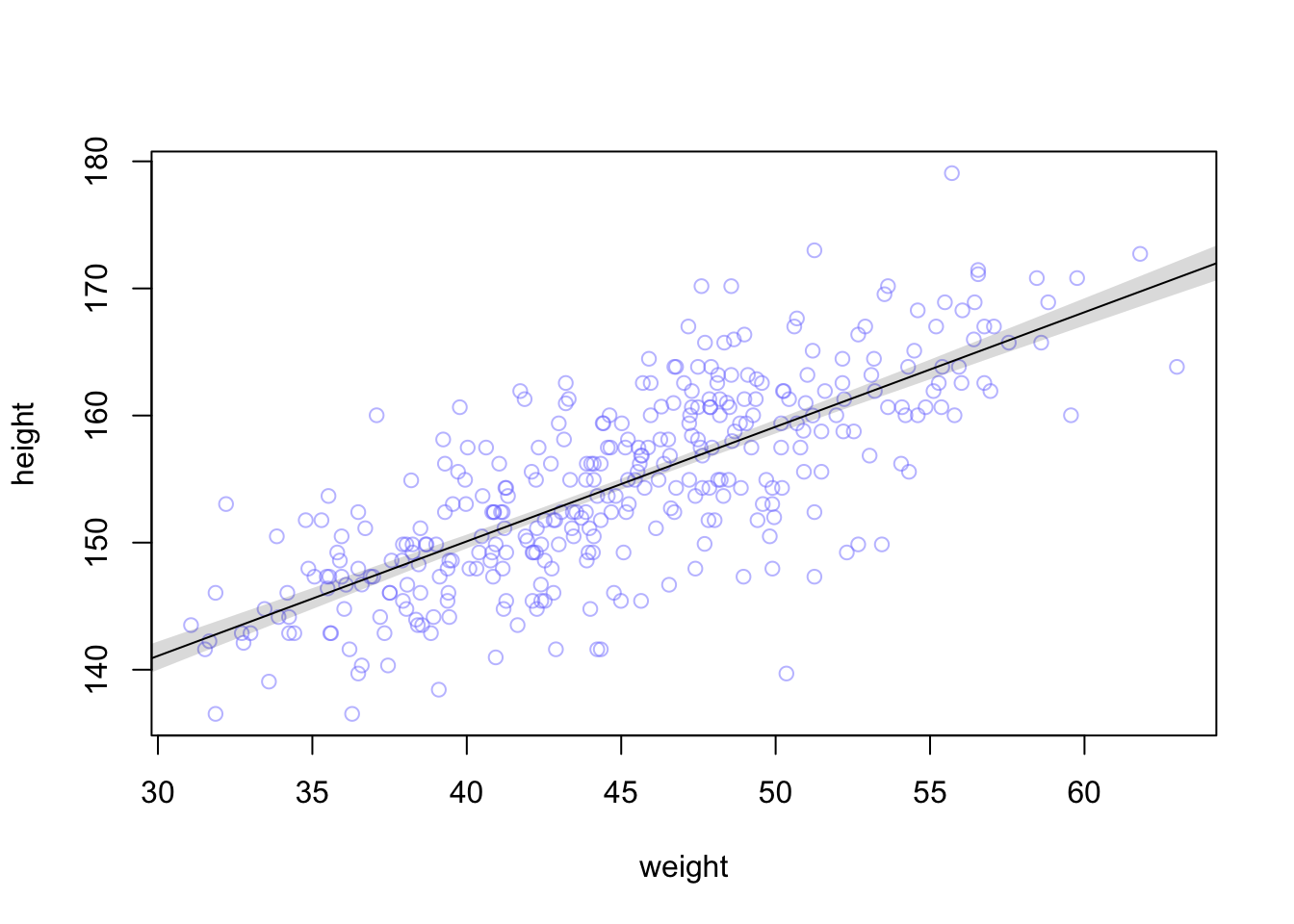
## integer(0)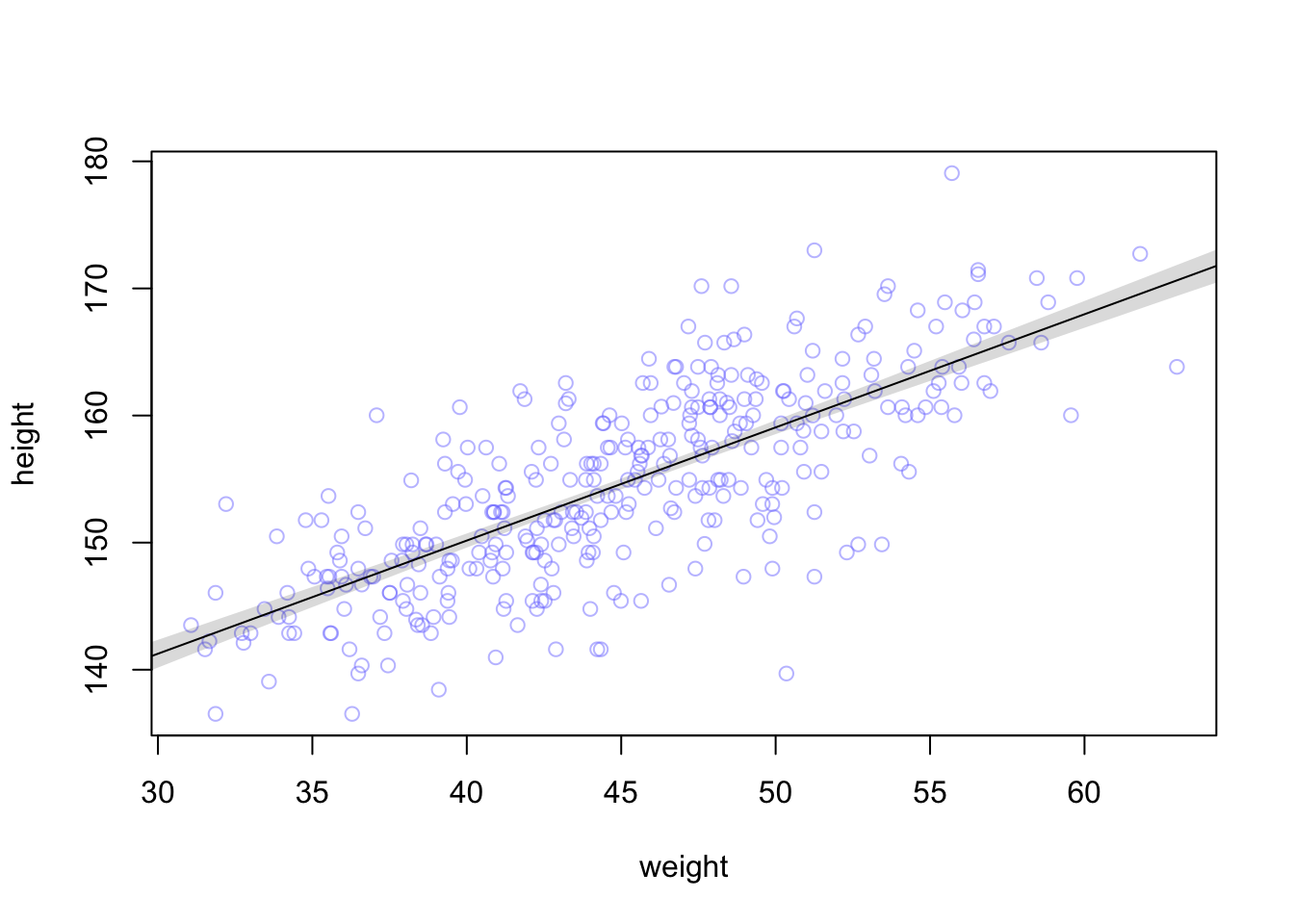
## integer(0)plot_grid(original, no_mean,labels = c("m4.3", "m4.3 - no mean"), ncol = 1)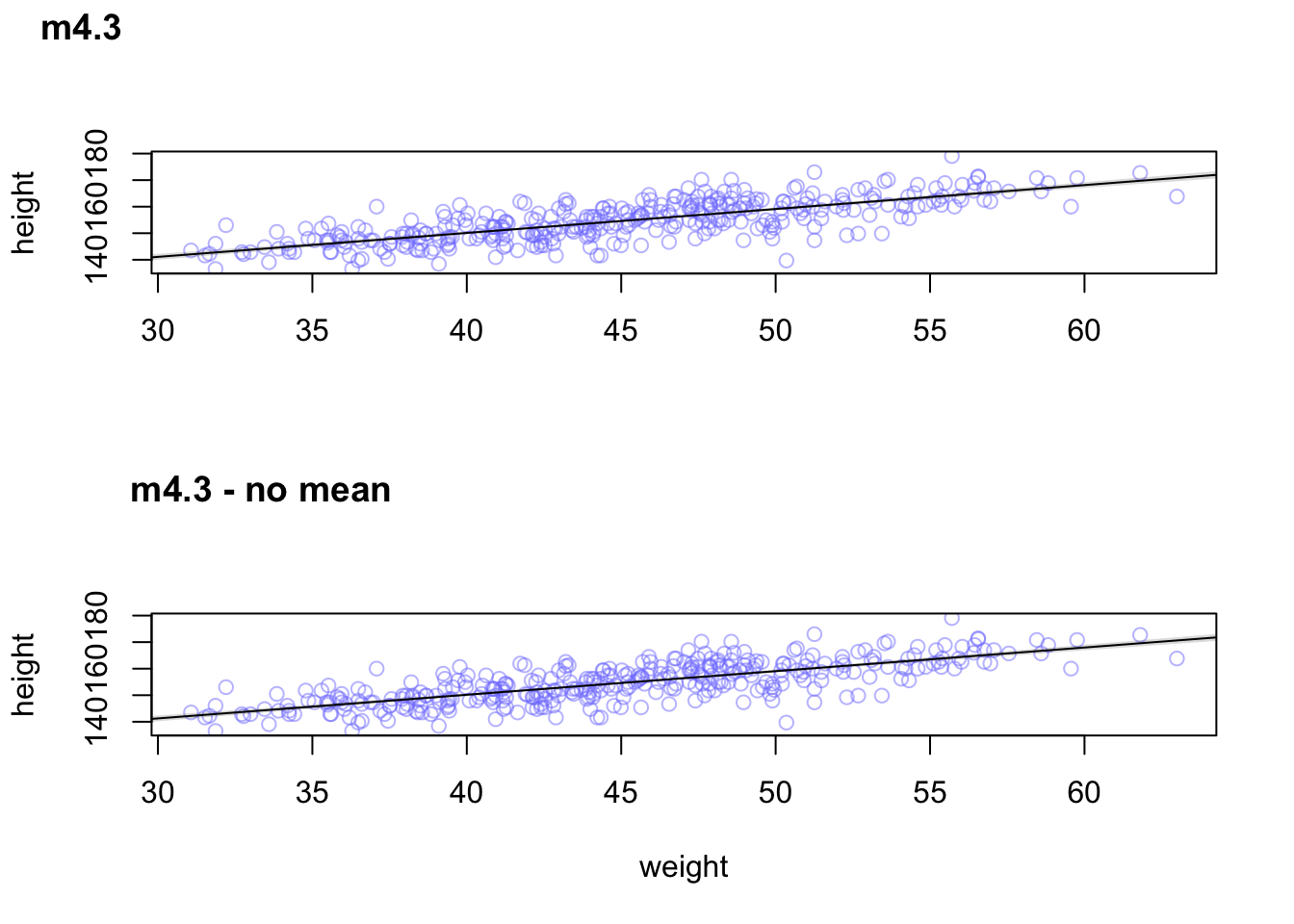
There’s no difference in the posterior predictions between the models.
4M8.
data("cherry_blossoms")
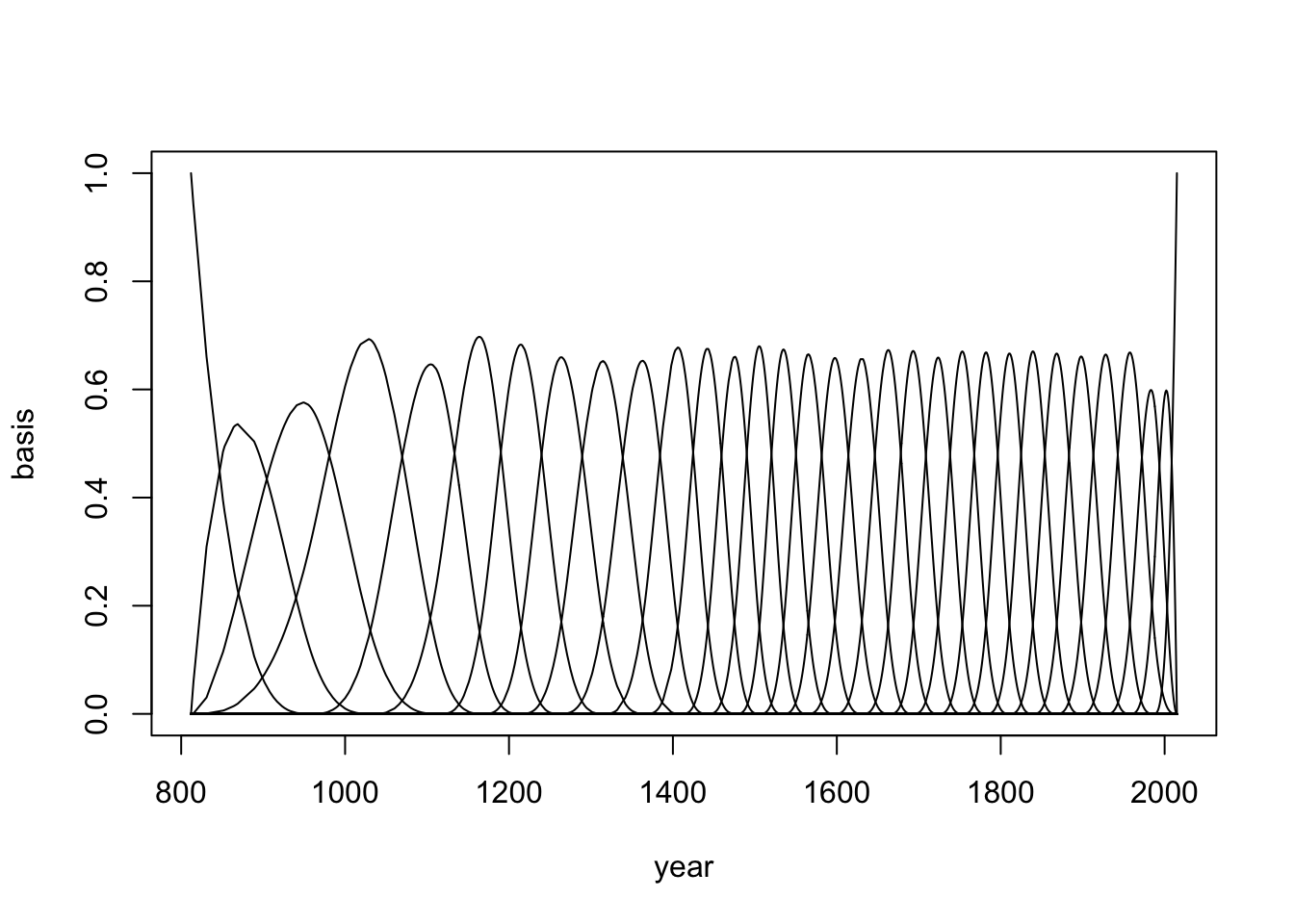
d <- cherry_blossoms15 Knots
20 Splines
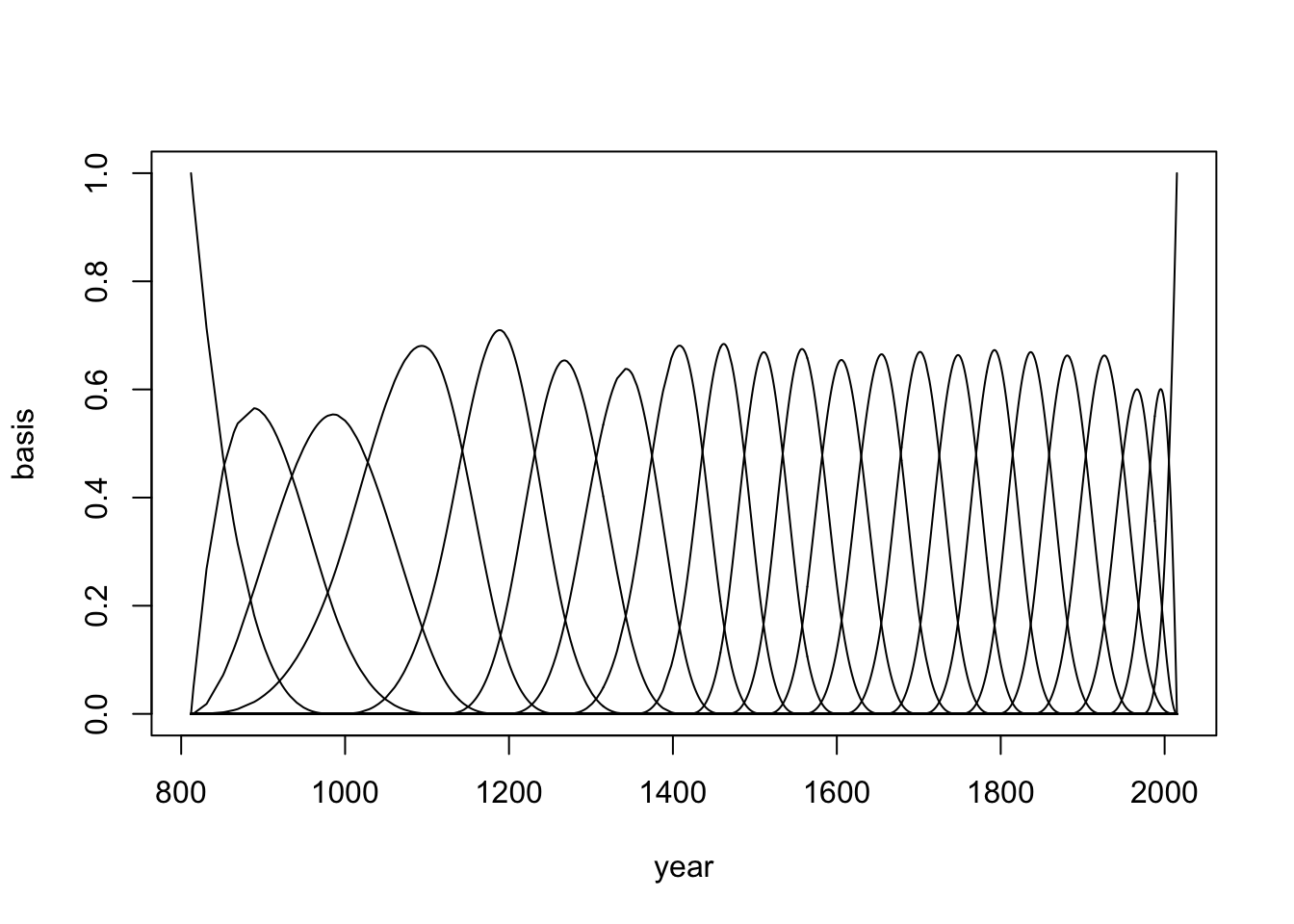 __ 30 Knots__
__ 30 Knots__